Ideje Rdkit Atom Index
Ideje Rdkit Atom Index. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Open source toolkit for cheminformatics Next, we initialized a feature vector, as described in yang et al. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
Nejchladnější Learn How To Make A Jupyter Notebook Widget For Annotation Of Atom Properties Cheminformania
Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Open source toolkit for cheminformatics Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Mar 01, 2021 · the rdkit documentation¶.The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Open source toolkit for cheminformatics Next, we initialized a feature vector, as described in yang et al. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:
This way the choice is independent of the atom order in the molecule... Open source toolkit for cheminformatics In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Next, we initialized a feature vector, as described in yang et al. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Mar 01, 2021 · the rdkit documentation¶. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.. This way the choice is independent of the atom order in the molecule.
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Open source toolkit for cheminformatics Mar 01, 2021 · the rdkit documentation¶. Next, we initialized a feature vector, as described in yang et al. (2019b), for each atom and bond based on computable features:.. This way the choice is independent of the atom order in the molecule.

In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant... An overview of the rdkit.
(2019b), for each atom and bond based on computable features: This way the choice is independent of the atom order in the molecule. Mar 01, 2021 · the rdkit documentation¶. Open source toolkit for cheminformatics. An overview of the rdkit.
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Open source toolkit for cheminformatics Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant... Next, we initialized a feature vector, as described in yang et al.. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:.. (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule... Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.. An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: (2019b), for each atom and bond based on computable features: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule. Open source toolkit for cheminformatics

Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule. An overview of the rdkit. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Open source toolkit for cheminformatics Next, we initialized a feature vector, as described in yang et al. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. (2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶... In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Next, we initialized a feature vector, as described in yang et al. Mar 01, 2021 · the rdkit documentation¶. An overview of the rdkit. (2019b), for each atom and bond based on computable features: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest... The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach... The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. (2019b), for each atom and bond based on computable features:. An overview of the rdkit.
An overview of the rdkit. An overview of the rdkit. Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Open source toolkit for cheminformatics This way the choice is independent of the atom order in the molecule. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Next, we initialized a feature vector, as described in yang et al.

An overview of the rdkit. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule. An overview of the rdkit... Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.
Open source toolkit for cheminformatics.. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: An overview of the rdkit.. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.

Next, we initialized a feature vector, as described in yang et al. Next, we initialized a feature vector, as described in yang et al. Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.. Open source toolkit for cheminformatics

Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,... When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. (2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. An overview of the rdkit. Next, we initialized a feature vector, as described in yang et al.. (2019b), for each atom and bond based on computable features:
Next, we initialized a feature vector, as described in yang et al... The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.

Mar 01, 2021 · the rdkit documentation¶.. Open source toolkit for cheminformatics An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest... In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
This way the choice is independent of the atom order in the molecule... Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Next, we initialized a feature vector, as described in yang et al. Mar 01, 2021 · the rdkit documentation¶. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. This way the choice is independent of the atom order in the molecule.

In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Open source toolkit for cheminformatics The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Next, we initialized a feature vector, as described in yang et al.
(2019b), for each atom and bond based on computable features:.. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Mar 01, 2021 · the rdkit documentation¶. Next, we initialized a feature vector, as described in yang et al. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Open source toolkit for cheminformatics When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: This way the choice is independent of the atom order in the molecule. (2019b), for each atom and bond based on computable features: When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:

In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. (2019b), for each atom and bond based on computable features: An overview of the rdkit. Next, we initialized a feature vector, as described in yang et al.. Next, we initialized a feature vector, as described in yang et al.
Open source toolkit for cheminformatics An overview of the rdkit. Mar 01, 2021 · the rdkit documentation¶. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Open source toolkit for cheminformatics Next, we initialized a feature vector, as described in yang et al. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
Open source toolkit for cheminformatics Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Mar 01, 2021 · the rdkit documentation¶. Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: This way the choice is independent of the atom order in the molecule. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.. Open source toolkit for cheminformatics
This way the choice is independent of the atom order in the molecule.. An overview of the rdkit. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Next, we initialized a feature vector, as described in yang et al. Open source toolkit for cheminformatics. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant.
Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.. Open source toolkit for cheminformatics Mar 01, 2021 · the rdkit documentation¶. Next, we initialized a feature vector, as described in yang et al. (2019b), for each atom and bond based on computable features: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. An overview of the rdkit. This way the choice is independent of the atom order in the molecule. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. An overview of the rdkit.
Mar 01, 2021 · the rdkit documentation¶. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. (2019b), for each atom and bond based on computable features: Open source toolkit for cheminformatics Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule. Mar 01, 2021 · the rdkit documentation¶. An overview of the rdkit. Next, we initialized a feature vector, as described in yang et al. Next, we initialized a feature vector, as described in yang et al.
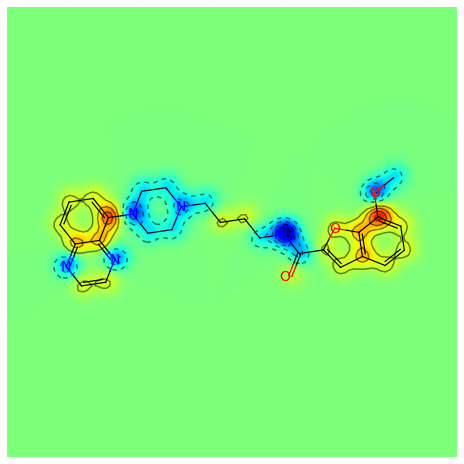
Mar 01, 2021 · the rdkit documentation¶... An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
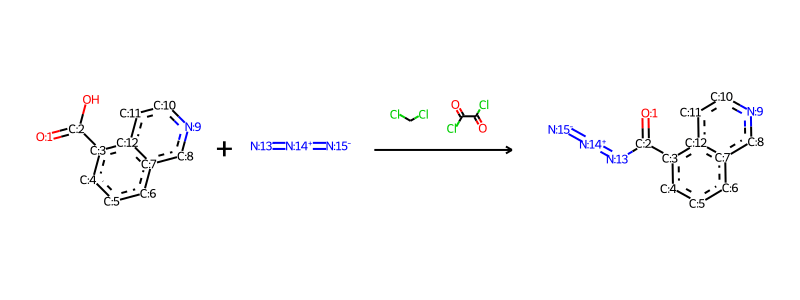
Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. . In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
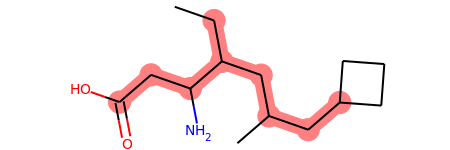
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant... . When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:
This way the choice is independent of the atom order in the molecule. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: An overview of the rdkit. Open source toolkit for cheminformatics Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features:.. An overview of the rdkit.
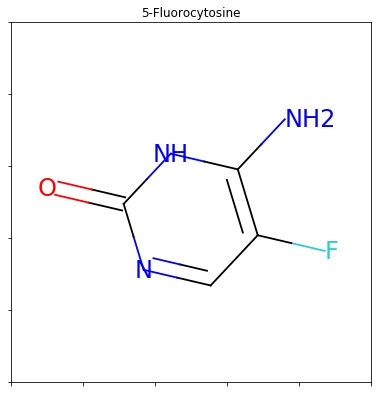
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: Next, we initialized a feature vector, as described in yang et al.. (2019b), for each atom and bond based on computable features:
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.. An overview of the rdkit.. An overview of the rdkit.
Open source toolkit for cheminformatics.. Mar 01, 2021 · the rdkit documentation¶. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Next, we initialized a feature vector, as described in yang et al. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. This way the choice is independent of the atom order in the molecule. Open source toolkit for cheminformatics An overview of the rdkit. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.
Mar 01, 2021 · the rdkit documentation¶. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. An overview of the rdkit. This way the choice is independent of the atom order in the molecule. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. (2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶... Open source toolkit for cheminformatics
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Mar 01, 2021 · the rdkit documentation¶. An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Open source toolkit for cheminformatics When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: (2019b), for each atom and bond based on computable features:

This way the choice is independent of the atom order in the molecule. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. An overview of the rdkit. Next, we initialized a feature vector, as described in yang et al. This way the choice is independent of the atom order in the molecule. Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:. Next, we initialized a feature vector, as described in yang et al.
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:.. Mar 01, 2021 · the rdkit documentation¶. Next, we initialized a feature vector, as described in yang et al.
Mar 01, 2021 · the rdkit documentation¶. Mar 01, 2021 · the rdkit documentation¶. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: This way the choice is independent of the atom order in the molecule. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Next, we initialized a feature vector, as described in yang et al. (2019b), for each atom and bond based on computable features:. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.

Next, we initialized a feature vector, as described in yang et al. Next, we initialized a feature vector, as described in yang et al. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.

Mar 01, 2021 · the rdkit documentation¶. Mar 01, 2021 · the rdkit documentation¶. Next, we initialized a feature vector, as described in yang et al. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.. Mar 01, 2021 · the rdkit documentation¶.
Next, we initialized a feature vector, as described in yang et al... Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: Open source toolkit for cheminformatics Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,... This way the choice is independent of the atom order in the molecule.

In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. An overview of the rdkit. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Next, we initialized a feature vector, as described in yang et al. (2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Mar 01, 2021 · the rdkit documentation¶.
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Open source toolkit for cheminformatics This way the choice is independent of the atom order in the molecule. Mar 01, 2021 · the rdkit documentation¶.
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:. This way the choice is independent of the atom order in the molecule. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
(2019b), for each atom and bond based on computable features:.. Next, we initialized a feature vector, as described in yang et al. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. This way the choice is independent of the atom order in the molecule. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Mar 01, 2021 · the rdkit documentation¶.. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach.. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant.
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:.. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. This way the choice is independent of the atom order in the molecule... (2019b), for each atom and bond based on computable features:
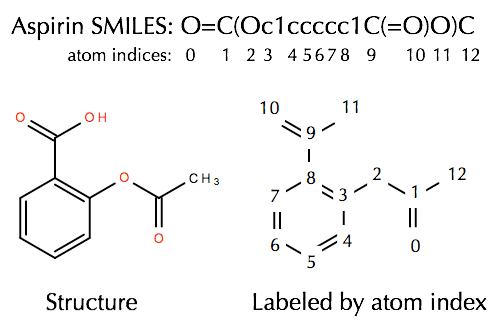
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Mar 01, 2021 · the rdkit documentation¶. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Next, we initialized a feature vector, as described in yang et al.. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:
Next, we initialized a feature vector, as described in yang et al.. Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. This way the choice is independent of the atom order in the molecule. Mar 01, 2021 · the rdkit documentation¶... This way the choice is independent of the atom order in the molecule.

The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest... This way the choice is independent of the atom order in the molecule. An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. Next, we initialized a feature vector, as described in yang et al. Open source toolkit for cheminformatics (2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant.
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule... The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.
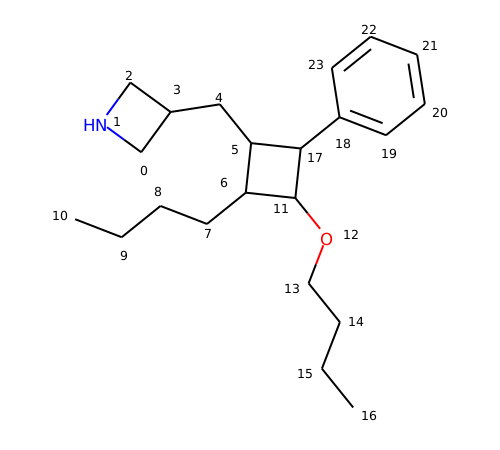
Open source toolkit for cheminformatics Open source toolkit for cheminformatics The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. This way the choice is independent of the atom order in the molecule. Next, we initialized a feature vector, as described in yang et al. Mar 01, 2021 · the rdkit documentation¶. An overview of the rdkit. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. (2019b), for each atom and bond based on computable features:
(2019b), for each atom and bond based on computable features: Mar 01, 2021 · the rdkit documentation¶. This way the choice is independent of the atom order in the molecule.

An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Next, we initialized a feature vector, as described in yang et al. Next, we initialized a feature vector, as described in yang et al.
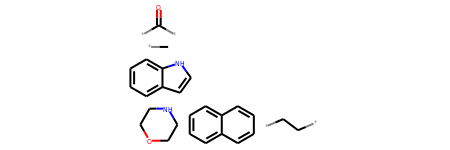
Mar 01, 2021 · the rdkit documentation¶. Mar 01, 2021 · the rdkit documentation¶. This way the choice is independent of the atom order in the molecule. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. (2019b), for each atom and bond based on computable features: An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Next, we initialized a feature vector, as described in yang et al. Open source toolkit for cheminformatics The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest.. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.
The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. Mar 01, 2021 · the rdkit documentation¶.. Open source toolkit for cheminformatics
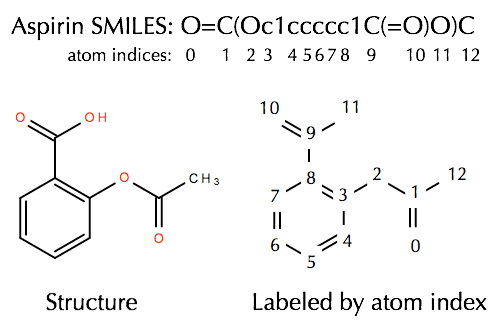
In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. This way the choice is independent of the atom order in the molecule. Next, we initialized a feature vector, as described in yang et al. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. Mar 01, 2021 · the rdkit documentation¶. (2019b), for each atom and bond based on computable features: Open source toolkit for cheminformatics When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Open source toolkit for cheminformatics

Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule. (2019b), for each atom and bond based on computable features: Open source toolkit for cheminformatics An overview of the rdkit. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:. (2019b), for each atom and bond based on computable features:

An overview of the rdkit.. Open source toolkit for cheminformatics In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Mar 01, 2021 · the rdkit documentation¶. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. (2019b), for each atom and bond based on computable features: Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. An overview of the rdkit. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. This way the choice is independent of the atom order in the molecule. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,.

Mar 01, 2021 · the rdkit documentation¶. Open source toolkit for cheminformatics This way the choice is independent of the atom order in the molecule. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. (2019b), for each atom and bond based on computable features: The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. An overview of the rdkit. Mar 01, 2021 · the rdkit documentation¶... (2019b), for each atom and bond based on computable features:
Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant.. Mar 01, 2021 · the rdkit documentation¶. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule. (2019b), for each atom and bond based on computable features: Next, we initialized a feature vector, as described in yang et al. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant.
This way the choice is independent of the atom order in the molecule. When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: An overview of the rdkit. Sep 01, 2019 · the rdkit implementation picks the atom with the smallest morgan invariant. This way the choice is independent of the atom order in the molecule. Jan 20, 2015 · 文章目录一、描述符计算模块1.rdkit.chem.lipinski模块2.rdkit.chem.descriptors模块3.rdkit.ml.descriptors.moleculedescriptors模块二、原子描述符可视化1.原子partial charge可视化2.原子logp可视化一、描述符计算模块1.rdkit.chem.lipinski模块rdkit中提供了许多描述符的计算方法,. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest... (2019b), for each atom and bond based on computable features:
When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract:.. In the case of symmetric atoms a and/or d, the rdkit implementation stores all possible torsional angles in the tf instead of only storing the smallest one as in the original approach. Next, we initialized a feature vector, as described in yang et al. The atom lists from getsubstructurematches are guaranteed to be in order of the smarts, but in this case we'll get five atoms so we need a way of picking out, in the correct order, the four of interest. (2019b), for each atom and bond based on computable features: When the smarts is parsed, the relevant atoms are assigned an atom map number property that we can easily extract: Open source toolkit for cheminformatics This way the choice is independent of the atom order in the molecule. Mar 01, 2021 · the rdkit documentation¶.. (2019b), for each atom and bond based on computable features:
